
Um\msports.h:46: ERROR_SUCCESS if the dialog was shown Um\MsiQuery.h:192:// Returns ERROR_SUCCESS if successful, and the view handle is returned, Um\Msi.h:1693:// Returns ERROR_SUCCESS if file is a package. Um\Mddefw.h:127: routine will return ERROR_SUCCESS and the inherited data even if &dwLcid, &dwSize) = ERROR_SUCCESS & dwType = REG_DWORD) || Um\MapiUnicodeHelp.h:583: if ((hkeyPolicy & RegQueryValueExW(hkeyPolicy, szName, 0, &dwType, (LPBYTE) Um\eappapis.h:56:// If the functions succeed, they return ERROR_SUCCESS. Um\eapmethodpeerapis.h:228:// If the function succeeds, it returns ERROR_SUCCESS.

Um\dsparse.h:102:_Success_(return = ERROR_SUCCESS) Um\ClusApi.h:571:_Success_(return = ERROR_SUCCESS) Um\advpub.h:40:// ERROR_SUCCESS_REBOOT_REQUIRED Reboot required. Shared\winerror.h:214:// MessageId: ERROR_SUCCESS
Shared\rpcnterr.h:34:#define RPC_S_OK ERROR_SUCCESS Shared\netioapi.h:2254: ERROR_SUCCESS on success.
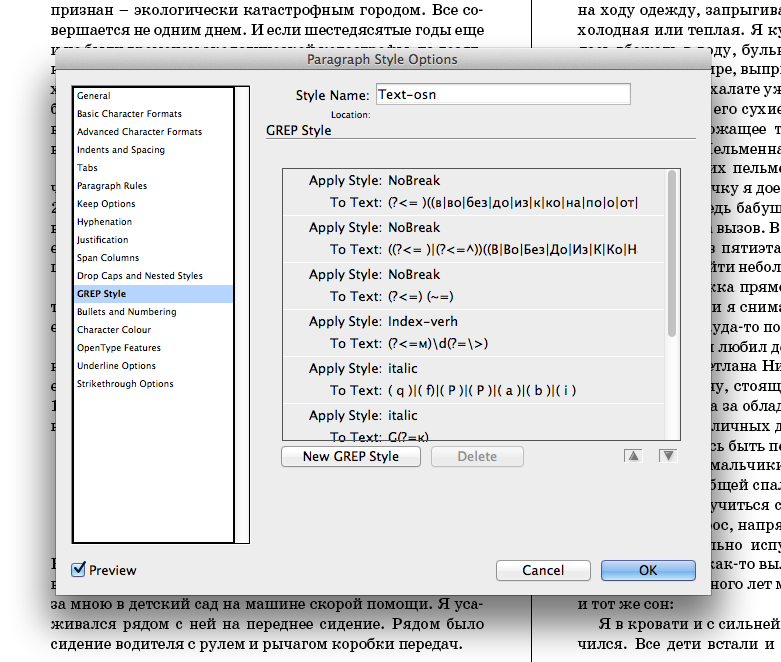
R GREP UNIQUE WINDOWS
Here's what some sample results look like (searching the Windows SDK for ERROR_SUCCESS): shared\bthdef.h:576:#define BTH_ERROR(_btStatus) ((_btStatus) != BTH_ERROR_SUCCESS)

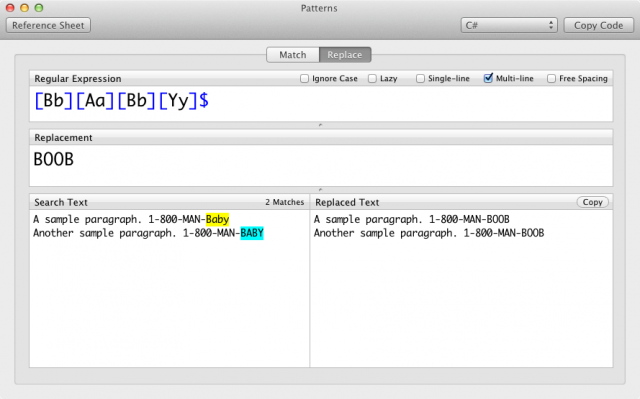
You can use it like this: gci -Recurse | sls -List FOOBAR Return only the first match in each input file.īy default, Select-String returns a MatchInfo object for each match found. Seurat v2.X vs v3.X Seurat v2.X Seurat v3.X GetAssayData(object = object) GetAssayData(object = object, slot = "counts") GetAssayData(object = object, slot = "scale.data") colnames(x = object) rownames(x = rownames(x = object) VariableFeatures(object = object) HVFInfo(object = object) object] object] GetCellEmbeddings(object = object, reduction.type = "pca") Embeddings(object = object, reduction = "pca") GetGeneLoadings(object = object, reduction.type = "pca") Loadings(object = object, reduction = "pca") AddMetaData(object = object, metadata = vector, col.Select-String has a -List parameter for this purpose: Seurat provides many prebuilt themes that can be added to ggplot2 plots for quick customization Theme Function DarkTheme Set a black background with white text FontSize Set font sizes for various elements of a plot NoAxes Remove axes and axis text NoLegend Remove all legend elements RestoreLegend Restores a legend after removal RotatedAxis Rotates x-axis labels # Dimensional reduction plot for PCA or tSNE DimPlot (object = pbmc, reduction = "tsne" ) DimPlot (object = pbmc, reduction = "pca" ) # Dimensional reduction plot, with cells colored by a quantitative feature FeaturePlot (object = pbmc, features = "MS4A1" ) # Scatter plot across single cells, replaces GenePlot FeatureScatter (object = pbmc, feature1 = "MS4A1", feature2 = "PC_1" ) FeatureScatter (object = pbmc, feature1 = "MS4A1", feature2 = "CD3D" ) # Scatter plot across individual features, repleaces CellPlot CellScatter (object = pbmc, cell1 = "AGTCTACTAGGGTG", cell2 = "CACAGATGGTTTCT" ) VariableFeaturePlot (object = pbmc ) # Violin and Ridge plots VlnPlot (object = pbmc, features = c ( "LYZ", "CCL5", "I元2" ) ) RidgePlot (object = pbmc, feature = c ( "LYZ", "CCL5", "I元2" ) ) # Heatmaps DoHeatmap (object = pbmc, features = heatmap_markers ) DimHeatmap (object = pbmc, reduction = "pca", cells = 200 ) # New things to try! Note that plotting functions now return ggplot2 objects, so you can add themes, titles, and # options onto them VlnPlot (object = pbmc, features = "MS4A1", = "groups" ) DotPlot (object = pbmc, features = c ( "LYZ", "CCL5", "I元2" ), = "groups" ) FeaturePlot (object = pbmc, features = c ( "MS4A1", "CD79A" ), blend = TRUE ) DimPlot (object = pbmc ) + DarkTheme ( ) DimPlot (object = pbmc ) + labs (title = "2,700 PBMCs clustered using Seurat and viewed\non a two-dimensional tSNE" )


 0 kommentar(er)
0 kommentar(er)
